Synthetic Biology
Much like Lego building blocks, genetic elements can be joined together to create new and interesting architecture, and the final construct can be transferred to a living organism which then executes the genetic instructions (DNA is transcribed into mRNA which is translated into protein).
One of my graduate school projects was to design and construct a genetic oscillator in mammalian cells, the result of which can be seen in the video below:
Here the individual cells produce and degrade a destabilized fluorescent protein which acts as the reporter of the state of the genetic circuit (tracked here by time-lapse fluorescence microscopy over the course of 92 hours).
The architecture of the genetic oscillator is shown below, where the various DNA elements are shown within the white boxes, while the regulatory mechanisms are depicted by orange arrows (activation) or purple bar-headed lines (inhibition).
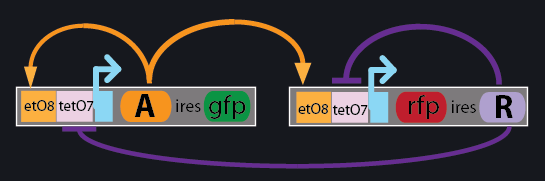
The oscillatory behavior is created by the negative feedback loop shown in purple, where the repressor protein R is expressed from a promoter that is also regulated by the very same repressor. Once the repressor protein reaches a sufficient threshold, it begins to shut down its own production. The activator protein also acts on the very same promoter in a positive feedback manner that quickly “fires” the system to a high-state, after which point the repressor shuts down the promoter until the proteins slowly degrade eventually reaching a threshold that allows the system to fire again and repeat this cycle.
Read more about the details of my design and the methodology in my dissertation by clicking on the PDF icon below.

